# Install and load libraries.
library(xlsx)
## Loading required package: rJava
## Loading required package: xlsxjars
library(ggplot2)
# Question 1
# Load Daphne Island data.
daphne.island <- read.xlsx("Homework 2 Data.xlsx", 1, header = FALSE)
daphne.island
## X1 X2 X3 X4 X5
## 1 8.65 10.61 10.89 8.42 9.73
## 2 9.05 8.94 11.29 9.52 8.47
## 3 10.52 10.25 9.55 8.62 9.68
## 4 8.89 10.25 9.24 11.53 9.59
## 5 8.67 9.98 10.54 10.26 9.43
# Load Santa Cruz data.
santa.cruz <- read.xlsx("Homework 2 Data.xlsx", 2, header = FALSE)
santa.cruz
## X1 X2 X3 X4 X5 X6 X7 X8 X9
## 1 12.66 12.35 10.88 11.29 8.47 10.52 11.01 10.46 11.78
## 2 11.04 11.56 10.33 10.62 10.43 9.74 10.02 10.91 10.30
## 3 10.82 11.07 10.21 10.49 12.09 13.25 8.73 10.27 11.82
## 4 13.49 10.65 10.68 9.99 10.24 9.47 10.18 9.81 NA
## 5 11.10 10.05 10.55 11.32 10.85 9.42 11.26 10.94 NA
# Display summaries for each island side-by-side; ignore 'NA' values where possible.
options(digits = 4)
finch.summary <- matrix(c((length(unlist(daphne.island)) - length(which(is.na(daphne.island)))),
(length(unlist(santa.cruz)) - length(which(is.na(santa.cruz)))),
mean(unlist(daphne.island)), mean(unlist(santa.cruz), na.rm = TRUE),
summary(unlist(daphne.island))[[3]], summary(unlist(santa.cruz))[[3]],
sd(unlist(daphne.island), na.rm = TRUE), sd(unlist(santa.cruz), na.rm = TRUE),
min(unlist(daphne.island), na.rm = TRUE), min(unlist(santa.cruz), na.rm = TRUE),
summary(unlist(daphne.island))[[2]], summary(unlist(santa.cruz))[[2]],
summary(unlist(daphne.island))[[5]], summary(unlist(santa.cruz))[[5]],
max(unlist(daphne.island), na.rm = TRUE), max(unlist(santa.cruz), na.rm = TRUE)),
ncol = 2, byrow = TRUE,
dimnames = list(c("Size", "Mean", "Median", "Std. Dev.", "Minimum", "First Quartile",
"Third Quartile", "Maximum"),
c("Daphne Island", "Santa Cruz")))
finch.summary
## Daphne Island Santa Cruz
## Size 25.0000 43.000
## Mean 9.7028 10.770
## Median 9.5900 10.650
## Std. Dev. 0.8894 1.024
## Minimum 8.4200 8.470
## First Quartile 8.9400 10.225
## Third Quartile 10.2600 11.180
## Maximum 11.5300 13.490
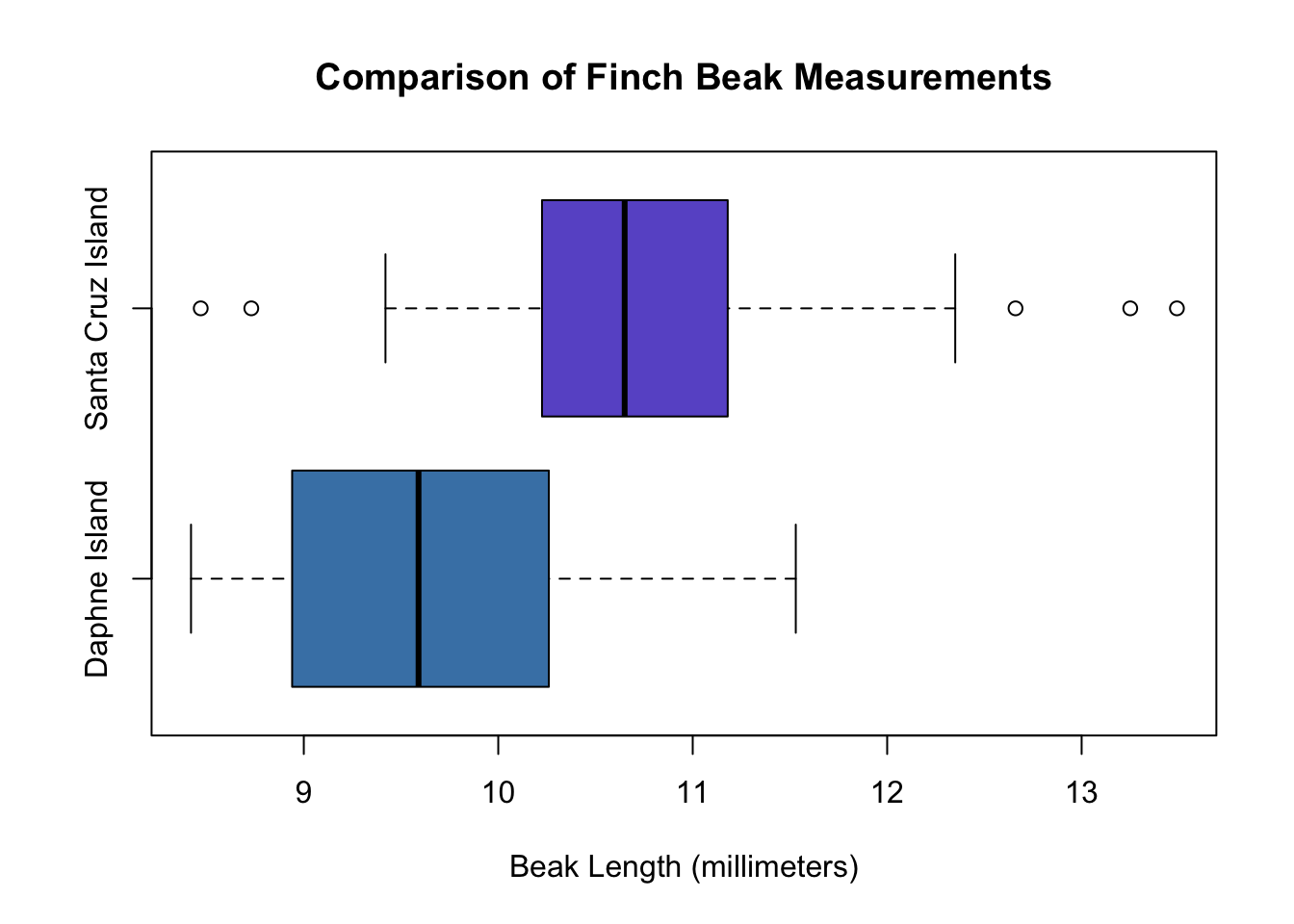
# Display graphs for each island's beak size distribution; use built-in boxplot function.
boxplot(unlist(daphne.island), unlist(santa.cruz), horizontal = TRUE,
main = "Comparison of Finch Beak Measurements",
names = c("Daphne Island", "Santa Cruz Island"),
xlab = "Beak Length (millimeters)",
col = c("steelblue", "slateblue"))
# Question 2
# Test if Daphne Island finch beak measurements differ from 10 mm.
t.test(unlist(daphne.island, use.names = FALSE), mu = 10, alternative = "two.sided")
##
## One Sample t-test
##
## data: unlist(daphne.island, use.names = FALSE)
## t = -1.7, df = 24, p-value = 0.1
## alternative hypothesis: true mean is not equal to 10
## 95 percent confidence interval:
## 9.336 10.070
## sample estimates:
## mean of x
## 9.703
# Question 3
# Find confidence interval of mean Daphne Island finch beak measurement using 90% confidence.
t.test(unlist(daphne.island, use.names = FALSE), conf.level = 0.90)
##
## One Sample t-test
##
## data: unlist(daphne.island, use.names = FALSE)
## t = 55, df = 24, p-value <2e-16
## alternative hypothesis: true mean is not equal to 0
## 90 percent confidence interval:
## 9.398 10.007
## sample estimates:
## mean of x
## 9.703
# Question 4
# Test if Daphne Island finch beak measurements are smaller than Santa Cruz Island finches.
t.test(unlist(daphne.island), unlist(santa.cruz), conf.level = 0.95, alternative = "less")
##
## Welch Two Sample t-test
##
## data: unlist(daphne.island) and unlist(santa.cruz)
## t = -4.5, df = 56, p-value = 2e-05
## alternative hypothesis: true difference in means is less than 0
## 95 percent confidence interval:
## -Inf -0.6716
## sample estimates:
## mean of x mean of y
## 9.703 10.770
